Our Work
We fund targeted efforts that remove bottlenecks — prototype builds, datasets, field infrastructure, and technical validation. Our goal is to help strong ideas become testable, publishable, and ready for follow-on support.
Tabula Madagascar
Tabula Madagascar is an ecosystem-scale genomics effort centered on Ranomafana National Park. The project builds the practical foundation needed to study biodiversity at molecular resolution—high-quality sample preservation, repeatable lab workflows, and the first wave of genomic and transcriptomic data across plants and arthropods that interact within the same ecosystem.
A core focus is getting the “field-to-data” pipeline right: standard operating procedures for collecting and processing samples for bulk DNA/RNA and single-cell workflows, and on-the-ground infrastructure and training at Centre ValBio so samples can be preserved and processed locally at high quality.
Eleftheria supports the work that turns collections into usable science: sample collection and documentation, preservation and extraction workflows, sequencing library construction and analysis, and the build-out of research capacity in Madagascar.

Dr. Stephen Quake
Despite the myriad advances in recent decades, we still lack clarity about many important molecular and cellular processes. Through this research, we hope to make great strides to preserve biodiversity by understanding the cellular complexity of key threatened organisms.
Rosetta: Solving the E. Coli Regulome
Rosetta is a large-scale effort to build a complete, predictive map of how E. coli genes are regulated at the promoter level. Despite decades of research, most E. coli promoters still lack reliable functional regulatory annotation. Rosetta addresses that gap by combining massively parallel genetics assays with systematic perturbations and modeling to identify regulatory elements, link them to transcription factors, and ultimately predict gene expression from DNA sequence and environmental context.
The goal is to create a base-pair–resolution atlas of regulatory DNA elements across the E. coli genome, establish causal “who binds where” links to transcription factors, and train a generative model that can both predict expression and help design new regulatory sequences.
Funding enables the scale: screening thousands of environmental and genetic conditions to activate otherwise silent promoters, building and testing genome-integrated promoter libraries, running deep sequencing at the required throughput, identifying transcription factors via CRISPRi and proteomics, and developing the computational infrastructure needed to assemble a public, usable atlas and predictive models.
Optical Imaging and Computation for White Blood Cell Counting
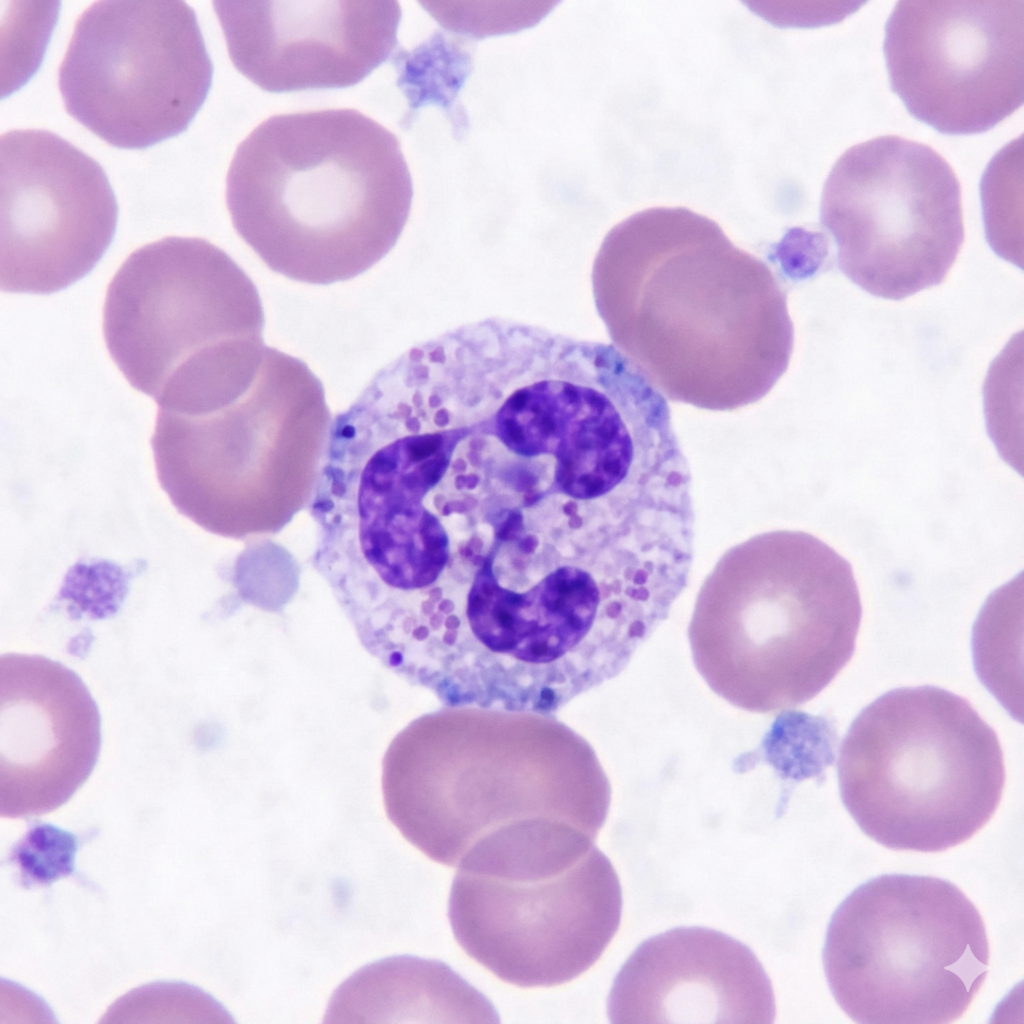
This project develops and tests a practical approach to counting white blood cells using optical imaging and computation. The work focuses on building a stroboscopic illumination setup synchronized to image acquisition, capturing long-exposure images, and developing a software pipeline to estimate white blood cell counts from those images.
Funding supports rapid prototyping (illumination control circuitry and synchronization), acquisition of test data, and development of the analysis repository and scripts needed to move from raw images to a repeatable counting readout, culminating in a concise technical summary of results and next steps.
